| Nature Science Update is an entertaining, informative and accessible round-up of what's new in the world of science, brought to you by the Nature News Service, the popular science syndication arm of the leading international science journal, Nature. |
| To find out about buying news and features like this for your website or news paper please e-mail:mailto:syndication@nature.com |
| Nature News Service celebrates the Human Genome |
| looking
back: How the sequence got the way it is |
| looking
on: Draft human genome published A journey through the genome: what's there Unfinished sympathy: what's missing? The Y chromosome: goldmine and junkyard |
| looking
forward: Biology goes back to the drawing board Ideas for a new biology Watching genes at work |
| Nature Genome Gateway >> |
Ideas for a new biology
PHILIP BALL
How will biologists construct a theory of the cell? Right now, the
field is wide open. But there is no shortage of ideas. Here are some of
them:
Networks
 |
Our societies trace out several vast webs, superimposed on one another. The information network of the World-Wide Web is just the most recent; there are also road, rail and air transportation networks, supply networks for electricity, fuel and water, networks of friendships and business contacts, and so on.
| the cell is a kind of community too |
The cell is a kind of community too, and relies on networks that have a surprising underlying similarity. If, for instance, every type of molecule in the cell is considered as a component of a network, and links are drawn between any two molecules that participate together in a biochemical process, the result is a network that has the same kind of connectivity structure as many social networks. The same may be true for networks that denote how one gene interacts with others in the genome.
This implies that cell biology has useful things to learn from mathematical network theory. Not all highly connected networks are the same. Some rapidly break down into isolated regions if only a few links are broken; others retain continuous pathways between almost any two nodes even with a high proportion of broken links.
The cell's network seems to be of this latter, robust variety—which makes good sense in evolutionary terms. By understanding the properties that a particular network shape confers, we can gain insights into how the cell organizes itself. Says Leroy Hood of the Institute for Systems Biology in Seattle, Washington, "the future will be the study of the genes and proteins of organisms in the context of their informational pathways and networks".
Modular biology
Genes often work in teams [see 'Watching genes at work']. When a cell breaks sugars down to harvest their energy, for instance, it might activate the whole team of genes to produce their respective enzymes.
Theoretical biologists Leland Hartwell, John Hopfield, Stan Leibler and Andrew Murray have suggested that we might be able to make sense of the buzzing activity in a cell by looking for examples of this kind of teamwork and considering them to represent more or less independent 'modules', like the separate departments of a large organization. One department takes care of making proteins, another replicates DNA ready for cell division, others receive and respond to particular hormones, and so on.
These modules are more than collections of genes: they include proteins, RNA, small messenger molecules and energy-rich molecules that together carry out a particular function.
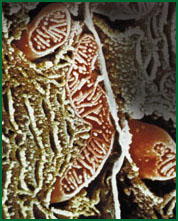 |
| A mitrochondrion, the cell organelle where energy production takes place. |
Sometimes the cell isolates such a module physically, enclosing it in a membrane-bound compartment, or 'organelle', like the mitochondrion, where energy production takes place. This is like giving the department its own building. Modules interact with one another via just one or a few types of mediating molecules, like departments that keep others informed of their activities with an occasional inter-office memo.
This approach avoids the need to account for everything at once. One could imagine devising a rather sophisticated model of the various interactions between the small number of molecules in a particular module, which would be far too complicated if extended to embrace the whole cell. The functioning of the module is then modified by a small number of 'inputs' from other modules, and produces only a small number of 'outputs' that affect other modules.
| rising above the details is an unfamiliar and uncomfortable notion for some molecular biologists; but they will need to get used to it |
Hartwell's group think that the idea of 'collective' behaviour familiar from physics might be useful for characterizing modules. "Most functional properties of a module are collective properties arising from the properties of the underlying components and their interactions. Collective properties have long been studied in physics and share attributes that rise above the details."
Rising above the details is an unfamiliar and uncomfortable notion for
some molecular biologists; but they will need to get used to
it.
Robustness
You can't expect things always to run smoothly in a system as complex as the cell. Little mistakes happen all the time. A protein is made with the wrong molecular structure, or in slightly the wrong amount. Raw ingredients are lacking if we haven't been eating a good diet. Toxins wander around causing havoc. Yet somehow life goes on, usually without our even noticing these problems. The cell is not a fragile thing.
| about 40% of our genes can be knocked out with no visible effect |
In recent years, geneticists have been surprised at just how robust it is. They might knock out a mouse gene thought to be crucial for survival, only to find that the mouse lives on. About 40% of our genes can be knocked out (individually) with no visible effect.
It has been proposed that this is because of gene redundancy: genes have almost identical 'back-up' copies that might switch on if the primary gene is switched off. But the latest research suggests that this is not the case: mutations in duplicated genes seem just as likely to have a physiological effect as those in unduplicated genes. So this robustness originates from some other feature of the genetic network.
It is already known that, for metabolic processes through which cells obtain energy and build new molecules, robustness can arise from 'module-wide' interactions: all the other metabolic genes adjust their activity to compensate for a faulty one. In other words, the robustness is not a property of any single molecule or gene but can be understood only at the collective level. Only recently was this metabolic phenomenon successfully described using mathematical control theory. Similar approaches may have to be developed for other, perhaps all, gene networks in the cell.
Another type of robustness is that, even if genes are working fine, cells sometimes seem not to care exactly how much of certain enzymes they have to hand, or how fast the enzymes work. In 1999 Stanislas Leibler at Princeton University and co-workers showed from experiments on bacteria that, again, this kind of robustness seems to stem from the 'nonlinear' properties of the biochemical pathway: the interactions and feedbacks between different protein enzymes.
"Robustness may be a common feature of many key cellular properties and
could be crucial for the reliable performance of many biochemical
networks," writes Leibler's team. One challenge will be to figure out
which features of a network are important in this context and which are
not.
Synthetic biology
If researchers were to create a theoretical or computer model of an entire cell, how would they know whether it was a good model? Physical scientists typically test their models against experiments by varying one quantity while keeping all the others constant, and by comparing model predictions with observations. In the cell that is extremely hard, if not impossible, to do; changing one thing almost inevitably changes others.
The answer might be to redesign cells to simplify them — just as, for example, an atmospheric physicist might test a theoretical model of planetary air circulation using a tank of fluid, since the experiment can't be conducted on the entire planet.
Some researchers have already created such tailor-made cells. Last year, Leibler's group came up with the idea of creating artificial networks of interacting genes that perform particular functions — for example switching the production of a protein on or off, or making its concentration oscillate. The team then genetically engineered Escherichia coli bacteria so that they contained these synthetic gene networks, and showed that they produced the predicted behaviour.
| scientists now believe they may be able to make very simple 'artificial cells' which can metabolize, replicate and evolve |
Even more ambitiously, Jack Szostak of the Howard Hughes Medical Institute in Boston, along with David Bartel of the Whitehead Institute in Massachusetts and Pier Luigi Luisi of the Eidgenössiche Technische Hochschule in Zürich, Switzerland, say that scientists now believe they may be able to make very simple 'artificial cells' from scratch which can metabolize, replicate and evolve. These cells would, in other words, fulfil the basic criteria for living entities while being wholly synthetic. This would mark "the beginning of the field of synthetic biology", according to Szostak and his colleagues.
Artificial cells, containing exactly what a researcher wanted to study, and nothing else, might then serve as miniature laboratories for probing models of molecular networks, and perhaps even for exploring how life became so complex in the first place.
© Macmillan Magazines Ltd 2001 - NATURE NEWS SERVICE
| Nature News Service celebrates the Human Genome |
| looking
back: How the sequence got the way it is |
| looking
on: Draft human genome published A journey through the genome: what's there Unfinished sympathy: what's missing? The Y chromosome: goldmine and junkyard |
| looking
forward: Biology goes back to the drawing board Ideas for a new biology Watching genes at work |
| Nature Genome Gateway >> |