Coding with Life's Code
Applications and developments in DNA-based computing |
By Aileen Constans
| Image: Courtesy of John Reif
|
 |
| |
The computer scientists, engineers, molecular and evolutionary biologists, and chemists involved in the relatively new field of DNA computing are taking this concept one step further. Ordinary biological processes, they say, are simply glorified computational algorithms, or sets of instructions for operating on data. But instead of crunching binary numbers (0 and 1), the DNA computer uses base-four math, as it were--A, C, G, and T. Consider a simple mathematical problem with 64 possible solutions. The entire solution set could be encoded in a set of three-base-long oligonucleotides, with AAA equaling 0, AAC equaling 1, AAG equaling 2, and so on. As the bioalgorithm runs--that is, as the pool of possible answers is subjected to a sequential series of enzymatic treatments--incorrect answers are discarded, leaving a set of solutions that researchers can then read.
The advantage of this approach is that while most computers test each possible solution one at a time, DNA computations execute in parallel--massively parallel. "In roughly one gram of DNA, you can have on the order of 1018 or more DNA strands, and in an experiment you can have each of these doing a basic operation," explains John Reif of Duke University.
Many potential applications exist, but desktop PCs are not likely to be replaced by test tubes full of biomolecular soup any time soon--if ever. "It's very important for people to realize that this capability probably isn't going to run Windows," cautions Reif. Nevertheless, researchers are forging ahead. Some scientists are combining DNA computing with evolutionary algorithms to make difficult-to-solve problems more tractable. Others are exploring how the self-assembly properties of DNA can be exploited in both computation and in the design of nanoscale structures or machines.
Only time will tell which of these approaches, if any, will ever transition from intellectual curiosity to actual tools scientists and engineers can use to solve real-world problems. Yet the nascent field of DNA computing is generating theories that could shape the studies of computing and of biological processes for years to come.
FLIGHT PLAN It began with a problem: Given a group of cities and their connecting flights, find if there exists a route between two endpoints that passes through the remaining cities exactly once. For a small number of cities, this problem, known as the directed Hamiltonian Path Problem (or the traveling-salesman problem [TSP]), is relatively simple to solve even without the aid of conventional computers. But as more cities are added it quickly becomes computationally intense. In the jargon of computer science, the TSP belongs to a class of problems called NP- complete. Such problems essentially require all possible solutions to be tested, making them more and more time-consuming as the number of variables increases.2
In a pioneering experiment, Leonard Adleman of the University of Southern California used DNA molecules to tackle a small (seven-city) version of the TSP. Adleman recognized parallels between a theoretical device called a Turing machine--a simplified computer that reads input data and uses it to write output data according to a set of instructions--and DNA polymerase. In this case, the input is a DNA strand. The polymerase, he observed, reads a series of bases off the template DNA, and according to the rules of Watson-Crick base pairing, writes the complementary string on a second, output strand.2
He synthesized oligonucleotides to represent each city and flight path between two cities. After allowing them to randomly combine into a set of possible answers, Adleman selected for paths that contained all the cities, began and ended with the desired cities, and were of the appropriate length. It took seven days, but the experiment worked.2
Critics were quick to point out potential problems. An order-of-magnitude scale-up of the experiment, for instance, would require a quantity of DNA greater than the Earth's mass.3 Nevertheless, computer scientists and molecular biologists alike were intrigued. Biochemical reactions are far slower than today's computer processors, but they also occur in parallel, which theoretically makes DNA-based computing ideal for solving highly complex problems that involve analyzing multiple solutions--problems that might take years to perform using even the most advanced conventional computer. "[Adleman's experiment] really opened up the field," says Harvey Rubin of the University of Pennsylvania.
| Image: Courtesy of John Reif |
 |
| |
SYSTEM OPTIMIZATION Several groups are now probing the power of DNA computing. In one experiment, Dirk Faulhammer and colleagues at Princeton University used a binary RNA library to solve a nine-variable problem, limiting error rates by employing a destructive algorithm in which RNA strands that represented incorrect answers were marked by complementary DNA strands for RNase H digestion.4
Robert Corn and colleagues at the University of Wisconsin tackled a four-variable problem using a DNA microarray-like approach: The set of possible solutions was attached to a solid support and subjected to rounds of hybridization and endonuclease digestion to yield the final answer.5 This approach, says Corn, offers several advantages over test tube-based methods. A chip format is more readily compatible with silicon devices, for instance, making it possible to couple DNA-chip technology with conventional computers in applications such as biodetection.
Last April, Adleman and colleagues solved a 20-variable, NP-complete problem with more than one million possible answers. The team encoded variables into short DNA "sticker" strands and employed oligonucleotide probes immobilized in a polyacrylamide gel to capture strands that contained complementary sequences. In this way, strands that encoded answers to the problem were selectively retained on the gel and could be separated and subjected to subsequent rounds of electrophoresis until a correct solution was obtained.6
Clearly, DNA computers have potential, but they will never overtake the power of conventional computers--which have solved a 13,509-city version of the TSP--without an influx of new ideas. A number of theorists have suggested incorporating evolutionary concepts, such as mutation, selection, and fitness crossover, to enrich the population of candidate answers for those that are most promising.4 David Harlan Wood of the University of Delaware adopted one such approach to simulate a DNA-based poker game. One player's DNA "strategies" compete against another's, and the most successful strategies are randomly modified, while unsuccessful ones are discarded. Successful strategies then compete with mutant strategies, and the process repeats until a winner emerges.7
| Image: Courtesy of John Reif |
 |
| |
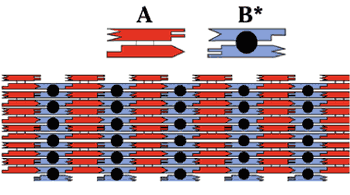
NANO-LEGO Another branch of the DNA-based computing field focuses on how DNA self-assembly can be used to orchestrate the assembly of nanoscale objects. Ned Seeman of New York University designs sequences that mimic Holliday junctions--naturally occurring branched motifs in which two double-stranded DNA molecules exchange strands, forming what looks like an intersection between two roads. Seeman's complexes contain a fixed branch point, and are capped with sticky ends so that the researchers can assemble relatively sophisticated structures from basic pieces--like molecular Lego.
Using these elements, the lab has created a variety of nanoscale structures, including cubes, octahedrons, and a simple DNA-based mechanical device that uses the B-Z transition of DNA as a "switch,"8 More recently, Bernard Yurke and colleagues at Bell Laboratories in Murray Hill, NJ, designed a tweezers-like assembly that uses DNA as a power source.8
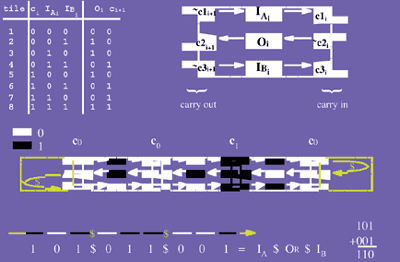
But how do these DNA structures and devices relate to DNA computing? Erik Winfree of the California Institute of Technology observed that Seeman's branched molecules resemble a theoretical construct called Wang tiles, square blocks with colored edges that associate as a mosaic according to a specific rule: Each edge must be surrounded by edges of the same color.9
Specific sets of Wang tiles can mimic the operation of a general-purpose computer. Seeman and collaborator Reif engineered branched DNA molecules that, when combined in a reaction tube, assembled like jigsaw puzzle pieces to perform a cumulative XOR operation--a binary computation that takes two input values and returns true if, and only if, the two inputs are different. In this case, instead of colors, each DNA tile possessed a unique set of sticky ends.8
This seemingly arcane exercise could have significant applications for both electronics and robotics. Reif views DNA nanofabrication as a potential substitute for lithography in the manufacture of extremely small computer chips. And Bruce Smith, president of Molecubotics, a start-up company that has collaborated with the Winfree lab, believes that DNA self-assembly is the fastest route to nanorobotics, as it provides a means to manufacture the molecular-scale building blocks that would be necessary to arrange nanoscale parts in an organized manner. "Pretty much any type of technology that moves into the nanoscale is going to need to rely on self-assembly to create structures," explains Winfree.
COMPUTING IN NATURE One of the most intriguing aspects of DNA computing research is the realization--perhaps not surprising--that even the simplest unicellular organisms conduct intricately complex molecular computations. Hypotrichous ciliates are single-celled organisms that contain two types of nuclei--a macronucleus that directs gene expression, and a micronucleus that is dormant until the cell reproduces. The arrangement of micronuclear genes precludes expression, as they are separated by short noncoding segments, and some of the separated genes are scrambled or encoded. But during reproduction, the genes are unscrambled and reassembled--in short, decoded--into the macronuclear form.
In 1999, Laura Landweber and Lila Kari at Princeton University recognized that this process closely resembled Adleman's approach to the Hamiltonian Path Problem, while Rozenberg and David Prescott of the University of Colorado, Boulder, determined that the splicing and linking procedure follows formal rules that parallel a linked list, which is a data structure commonly used in conventional computing.10
Now researchers hope to harness these processes to perform computations of their own design. University of Pennsylvania's Rubin says DNA could be introduced into unicellular organisms such as bacteria to allow them to sense an element in the environment and "compute" a response, such as slowing growth or increasing expression of a particular protein. "We might be able to engineer cells to respond to their environment in a particular way," he says.
Engineered cells could then be combined with recording devices to build sensors similar to today's reporter-based detection systems, but with a difference: The cell would be able to fine-tune the level of detection sensitivity based on what it senses in the environment. "I can put a receptor on the surface of a bacteria and then hook that up to a reporter molecule that glows. That's done all the time," Rubin explains. "What's not done all the time is to actually program that in the sense of being able to control the level of glowing or the level of sensitivity, or to have the bugs do something more than glow." Rubin emphasizes, however, that this type of technology remains to be developed.
For DNA-based computing, therein lies the rub: Most of the existing applications remain theoretical possibilities, and it is difficult to tell what the future holds. "DNA computation is sort of like aviation in about 1905. There was such a thing as an airplane, but who knew if it was actually going to become a major mode of transportation or just sort of a toy?" asks Seeman.
Researchers in the field are treading cautiously in these uncharted waters, but most agree that the cooperation between the two seemingly disparate disciplines of computer science and bioscience will lead to important new insights in both fields. "I think that in maybe 20 years we will have a science of computation that is dramatically different from what we know about computation today," says Rozenberg. "Even if we don't get anything very specific [in terms of practical applications], the understanding of what computation is will become deeper and much broader."
Aileen Constans (aconstans@the-scientist.com) is a contributing editor.
References
1. D.R. Hofstadter, Gödel, Escher, Bach: An Eternal Golden Braid,
New York: Vintage Books, 1979.
2. L.M. Adleman, "Molecular computation of solutions to combinatorial
problems," Science, 266:1021-4, 1994.
3. M. Linial, N. Linial, "On the potential of molecular computing,"
Science, 268:481, 1995.
4.. J. Chen, D.H. Wood, "Computation with biomolecules," Proceedings
of the National Academy of Sciences, 97:1328-30, 2000.
5. Q. Liu et al., "DNA computing on surfaces," Nature, 403:175-9,
2000.
6. R.S. Braich et al., "Solution of a 20-variable 3-SAT problem on a
DNA computer," Science, 296:499-502, April 19, 2002.
7. D.H. Wood et al., "DNA starts to learn to play poker," in DNA
Computing, 7th International Workshop on DNA-Based Computers, N.
Jonoska, N.C. Seeman, eds., New York: Springer-Verlag, 2002, pp. 92-103.
8. J. Reif, "The emergence of the discipline of biomolecular computing
in the US," New Generation Computing, 20:1-23, 2002
9. E. Winfree, "Algorithmic self-assembly of DNA: Theoretical motivations
and 2D assembly experiments," Journal of Biomolecular Structure and
Dynamics, 2:263-70, 2000.
10. G. Rozenberg, "Gene assembly in ciliates: Computing by folding and
recombination," in A Half-Century of Automata Theory - Celebration
and Inspiration, A. Salomaa et al., eds., River Edge, NJ: World
Scientific, 2001, pp. 93-130.